Publications
2025
- Tales of Morality: Comparing Human- and LLM-Generated Moral Stories from Visual CuesRezvaneh Rezapour, Sullam Jeoung, Zhiwen You, and Jana DiesnerIn Findings of the Association for Computational Linguistics: EMNLP 2025, Nov 2025
Do moral values align between images, the stories humans write about them, and the narratives generated by large language models (LLMs)? This question matters because stories are central to how humans communicate moral values, yet little is known about how people and LLMs perform this task in a multimodal (text and image) setting. We present a systematic comparison of moral values represented in human- and LLM-generated narratives based on images annotated by humans for moral content. Our analysis shows that while human stories reflect a balanced distribution of moral foundations and coherent narrative arcs, LLMs disproportionately emphasize the Care foundation and often lack emotional resolution. Even with moral conditioning, these biases persist in LLMs. We introduce a novel dataset and framework for evaluating moral storytelling in vision-language models, highlighting key challenges in aligning AI with human moral reasoning across cultures.
-
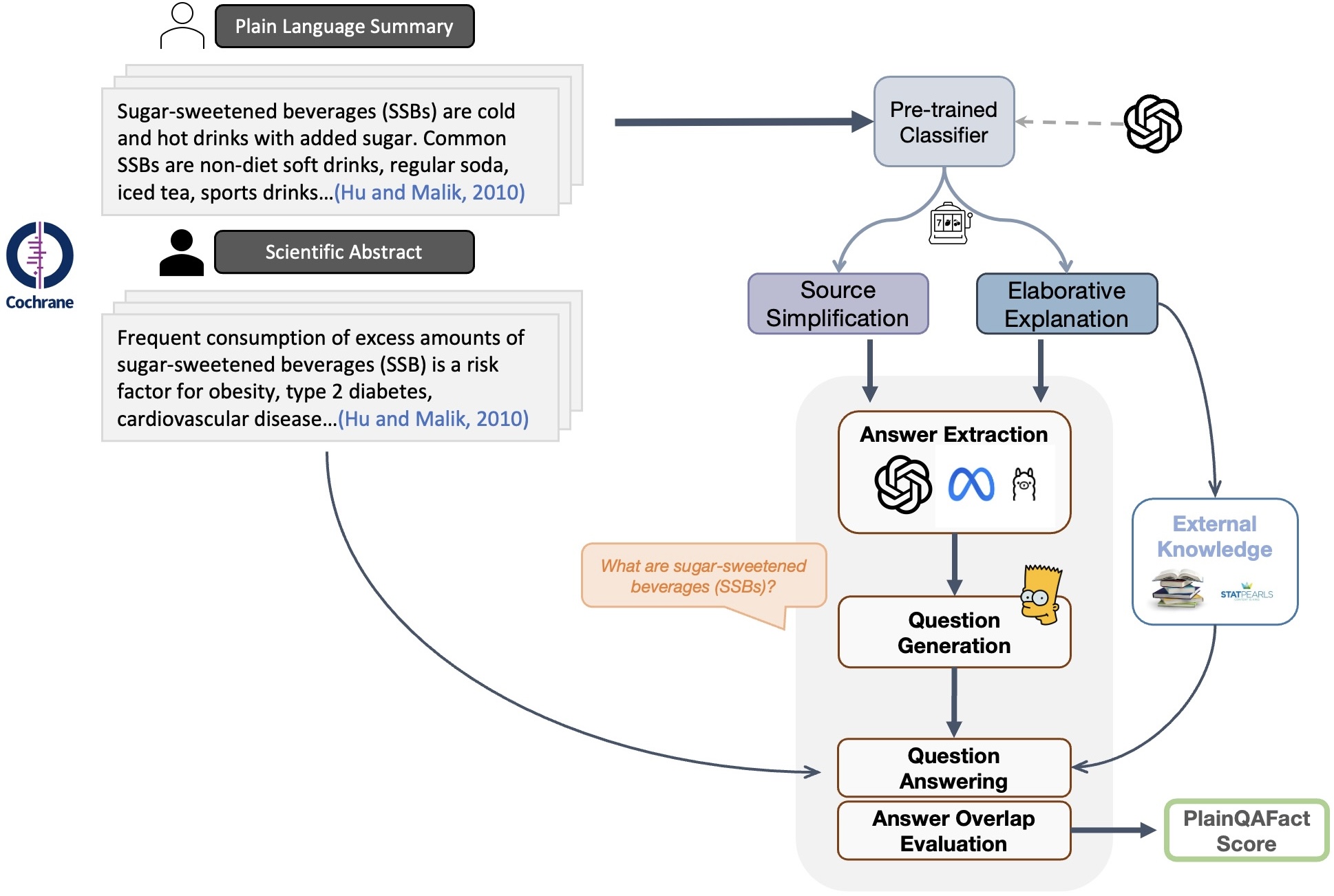 PlainQAFact: Retrieval-augmented Factual Consistency Evaluation Metric for Biomedical Plain Language SummarizationZhiwen You, and Yue GuoarXiv preprint arXiv:2503.08890, Nov 2025
PlainQAFact: Retrieval-augmented Factual Consistency Evaluation Metric for Biomedical Plain Language SummarizationZhiwen You, and Yue GuoarXiv preprint arXiv:2503.08890, Nov 2025Hallucinated outputs from large language models (LLMs) pose risks in the medical domain, especially for lay audiences making health-related decisions. Existing automatic factual consistency evaluation methods, such as entailment- and question-answering (QA) -based, struggle with plain language summarization (PLS) due to elaborative explanation phenomenon, which introduces external content (e.g., definitions, background, examples) absent from the scientific abstract to enhance comprehension. To address this, we introduce PlainQAFact, an automatic factual consistency evaluation metric trained on a fine-grained, human-annotated dataset PlainFact, for evaluating factual consistency of both source-simplified and elaborately explained sentences. PlainQAFact first classifies sentence type, then applies a retrieval-augmented QA scoring method. Empirical results show that existing evaluation metrics fail to evaluate the factual consistency in PLS, especially for elaborative explanations, whereas PlainQAFact consistently outperforms them across all evaluation settings. We further analyze PlainQAFact’s effectiveness across external knowledge sources, answer extraction strategies, answer overlap measures, and document granularity levels, refining its overall factual consistency assessment. Taken together, our work presents the first evaluation metric designed for PLS factual consistency evaluation, providing the community with both a robust benchmark and a practical tool to advance reliable and safe plain language communication in the medical domain.
- Revisiting gender bias research in bibliometrics: Standardizing methodological variability using Scholarly Data Analysis (SoDA) CardsHaeJin Lee, Shubhanshu Mishra, Apratim Mishra, Zhiwen You, Jinseok Kim, and Jana DiesnerarXiv preprint arXiv:2501.18129, Nov 2025
Gender biases in scholarly metrics remain a persistent concern, despite numerous bibliometric studies exploring their presence and absence across productivity, impact, acknowledgment, and self-citations. However, methodological inconsistencies, particularly in author name disambiguation and gender identification, limit the reliability and comparability of these studies, potentially perpetuating misperceptions and hindering effective interventions. A review of 70 relevant publications over the past 12 years reveals a wide range of approaches, from name-based and manual searches to more algorithmic and gold-standard methods, with no clear consensus on best practices. This variability, compounded by challenges such as accurately disambiguating Asian names and managing unassigned gender labels, underscores the urgent need for standardized and robust methodologies. To address this critical gap, we propose the development and implementation of "Scholarly Data Analysis (SoDA) Cards." These cards will provide a structured framework for documenting and reporting key methodological choices in scholarly data analysis, including author name disambiguation and gender identification procedures. By promoting transparency and reproducibility, SoDA Cards will facilitate more accurate comparisons and aggregations of research findings, ultimately supporting evidence-informed policymaking and enabling the longitudinal tracking of analytical approaches in the study of gender and other social biases in academia.
2024
-
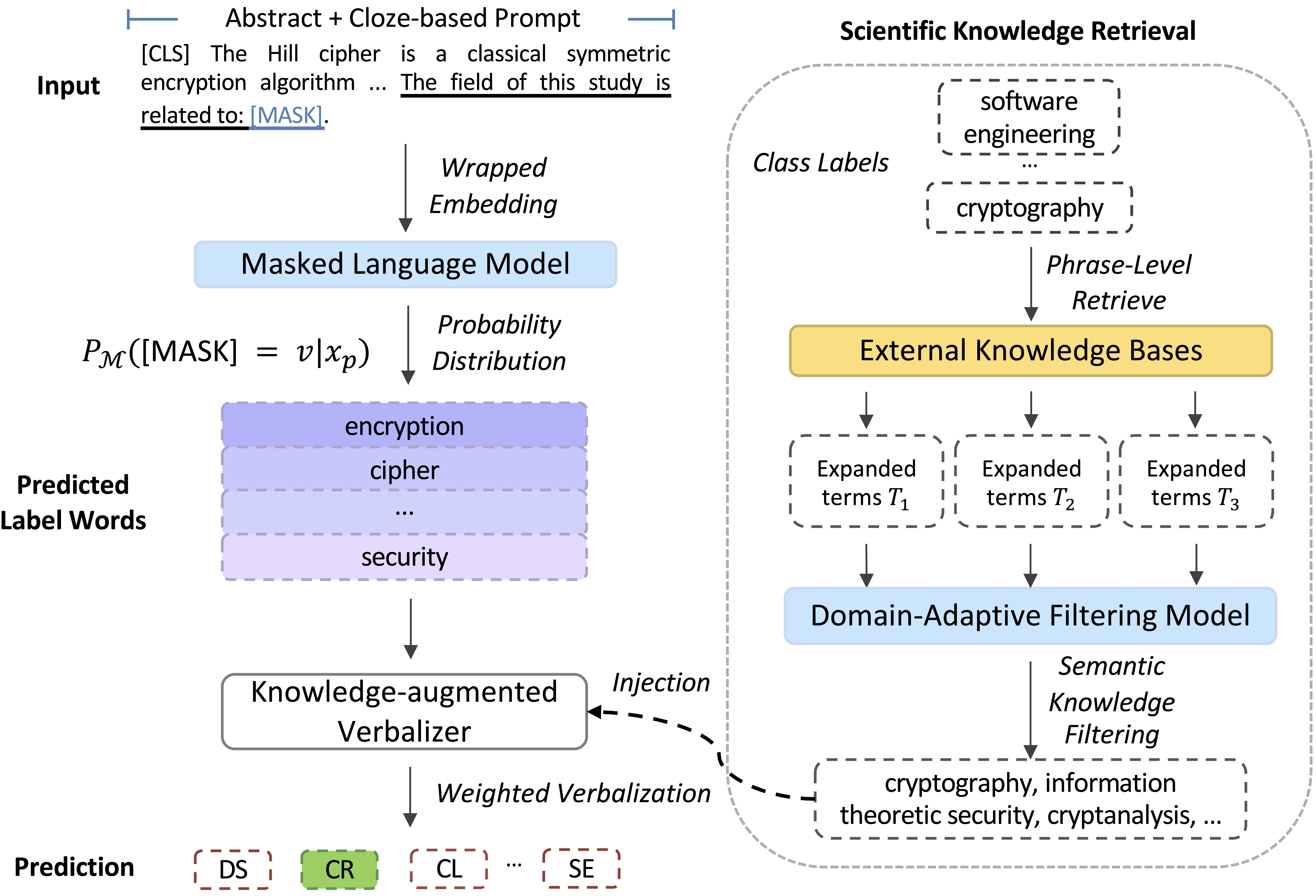 SciPrompt: Knowledge-augmented Prompting for Fine-grained Categorization of Scientific TopicsZhiwen You, Kanyao Han, Haotian Zhu, Bertram Ludaescher, and Jana DiesnerIn Proceedings of the 2024 Conference on Empirical Methods in Natural Language Processing, Nov 2024
SciPrompt: Knowledge-augmented Prompting for Fine-grained Categorization of Scientific TopicsZhiwen You, Kanyao Han, Haotian Zhu, Bertram Ludaescher, and Jana DiesnerIn Proceedings of the 2024 Conference on Empirical Methods in Natural Language Processing, Nov 2024Prompt-based fine-tuning has become an essential method for eliciting information encoded in pre-trained language models for a variety of tasks, including text classification. For multi-class classification tasks, prompt-based fine-tuning under low-resource scenarios has resulted in performance levels comparable to those of fully fine-tuning methods. Previous studies have used crafted prompt templates and verbalizers, mapping from the label terms space to the class space, to solve the classification problem as a masked language modeling task. However, cross-domain and fine-grained prompt-based fine-tuning with an automatically enriched verbalizer remains unexplored, mainly due to the difficulty and costs of manually selecting domain label terms for the verbalizer, which requires humans with domain expertise. To address this challenge, we introduce SciPrompt, a framework designed to automatically retrieve scientific topic-related terms for low-resource text classification tasks. To this end, we select semantically correlated and domain-specific label terms within the context of scientific literature for verbalizer augmentation. Furthermore, we propose a new verbalization strategy that uses correlation scores as additional weights to enhance the prediction performance of the language model during model tuning. Our method outperforms state-of-the-art, prompt-based fine-tuning methods on scientific text classification tasks under few and zero-shot settings, especially in classifying fine-grained and emerging scientific topics.
-
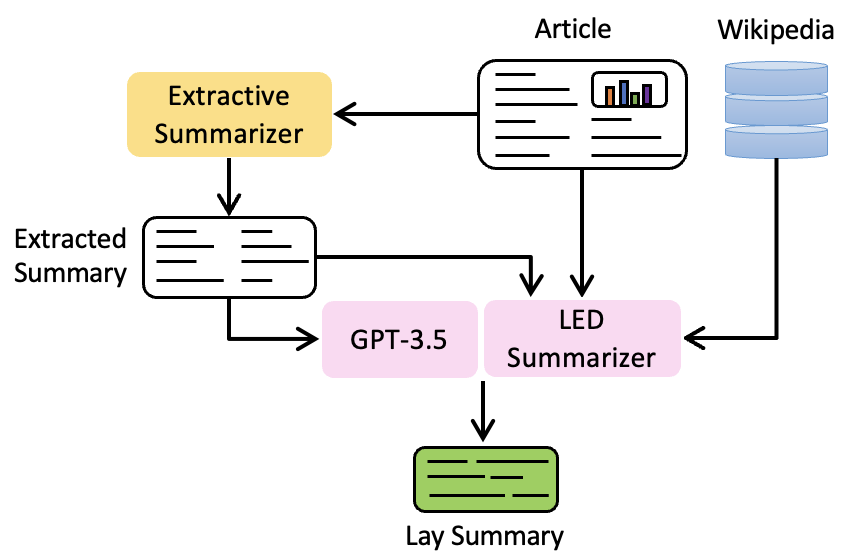 UIUC_BioNLP at BioLaySumm: An Extract-then-Summarize Approach Augmented with Wikipedia Knowledge for Biomedical Lay SummarizationZhiwen You, Shruthan Radhakrishna, Shufan Ming, and Halil KilicogluIn Proceedings of the 23rd Workshop on Biomedical Natural Language Processing, Aug 2024
UIUC_BioNLP at BioLaySumm: An Extract-then-Summarize Approach Augmented with Wikipedia Knowledge for Biomedical Lay SummarizationZhiwen You, Shruthan Radhakrishna, Shufan Ming, and Halil KilicogluIn Proceedings of the 23rd Workshop on Biomedical Natural Language Processing, Aug 2024As the number of scientific publications is growing at a rapid pace, it is difficult for laypeople to keep track of and understand the latest scientific advances, especially in the biomedical domain. While the summarization of scientific publications has been widely studied, research on summarization targeting laypeople has remained scarce. In this study, considering the lengthy input of biomedical articles, we have developed a lay summarization system through an extract-then-summarize framework with large language models (LLMs) to summarize biomedical articles for laypeople. Using a fine-tuned GPT-3.5 model, our approach achieves the highest overall ranking and demonstrates the best relevance performance in the BioLaySumm 2024 shared task.
-
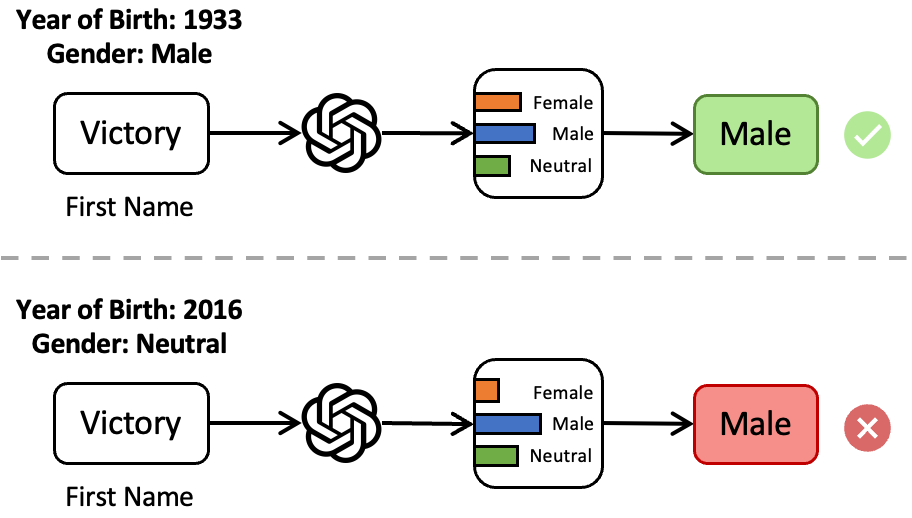 Beyond Binary Gender Labels: Revealing Gender Bias in LLMs through Gender-Neutral Name PredictionsZhiwen You, HaeJin Lee, Shubhanshu Mishra, Sullam Jeoung, Apratim Mishra, Jinseok Kim, and Jana DiesnerIn Proceedings of the 5th Workshop on Gender Bias in Natural Language Processing (GeBNLP), Aug 2024
Beyond Binary Gender Labels: Revealing Gender Bias in LLMs through Gender-Neutral Name PredictionsZhiwen You, HaeJin Lee, Shubhanshu Mishra, Sullam Jeoung, Apratim Mishra, Jinseok Kim, and Jana DiesnerIn Proceedings of the 5th Workshop on Gender Bias in Natural Language Processing (GeBNLP), Aug 2024Name-based gender prediction has traditionally categorized individuals as either female or male based on their names, using a binary classification system. That binary approach can be problematic in the cases of gender-neutral names that do not align with any one gender, among other reasons. Relying solely on binary gender categories without recognizing gender-neutral names can reduce the inclusiveness of gender prediction tasks. We introduce an additional gender category, i.e., "neutral", to study and address potential gender biases in Large Language Models (LLMs). We evaluate the performance of several foundational and large language models in predicting gender based on first names only. Additionally, we investigate the impact of adding birth years to enhance the accuracy of gender prediction, accounting for shifting associations between names and genders over time. Our findings indicate that most LLMs identify male and female names with high accuracy (over 80%) but struggle with gender-neutral names (under 40%), and the accuracy of gender prediction is higher for English-based first names than non-English names. The experimental results show that incorporating the birth year does not improve the overall accuracy of gender prediction, especially for names with evolving gender associations. We recommend using caution when applying LLMs for gender identification in downstream tasks, particularly when dealing with non-binary gender labels.